4.2.4. Parallel analysis
Added in version 2.8.0: Parallelization of analysis classes was added during Google Summer of Code 2023 by @marinegor and MDAnalysis GSoC mentors.
This section explains the implementation and background for
parallelization with the MDAnalysis.analysis.base.AnalysisBase, what
users and developers need to know, when you should use parallelization (almost
always!), and when you should abstain from doing so (rarely).
4.2.4.1. How to use parallelization
In order to use parallelization in a built-in analysis class SomeAnalysisClass,
simply check which backends are available (see Backends for backends
that are generally available), and then just enable them by providing
backend='multiprocessing' and n_workers=... to SomeClass.run(...)
method:
u = mda.Universe(...)
my_run = SomeClass(trajectory)
assert SomeClass.get_supported_backends() == ('serial', 'multiprocessing', 'dask')
my_run.run(backend='multiprocessing', n_workers=12)
For some classes, such as MDAnalysis.analysis.rms.RMSF (in its current implementation),
split-apply-combine parallelization isn’t possible, and running them will be
impossible with any but the serial backend.
Note
Parallelization is getting added to existing analysis classes. Initially,
only MDAnalysis.analysis.rms.RMSD supports parallel analysis, but
we aim to increase support in future releases. Please check issues labeled
parallelization on the MDAnalysis issues tracker.
4.2.4.2. How does parallelization work
The main idea behind its current implementation is that a trajectory analysis is often trivially parallelizable, meaning you can analyze all frames independently, and then merge them in a single object. This approach is also known as “split-apply-combine”, and isn’t new to MDAnalysis users, since it was first introduced in PMDA [1]. Version 2.8.0 of MDAnalysis brings this approach to the main library.
4.2.4.2.1. split-apply-combine
The following scheme explains the current AnalysisBase.run protocol (user-implemented methods
are highlighted in orange):
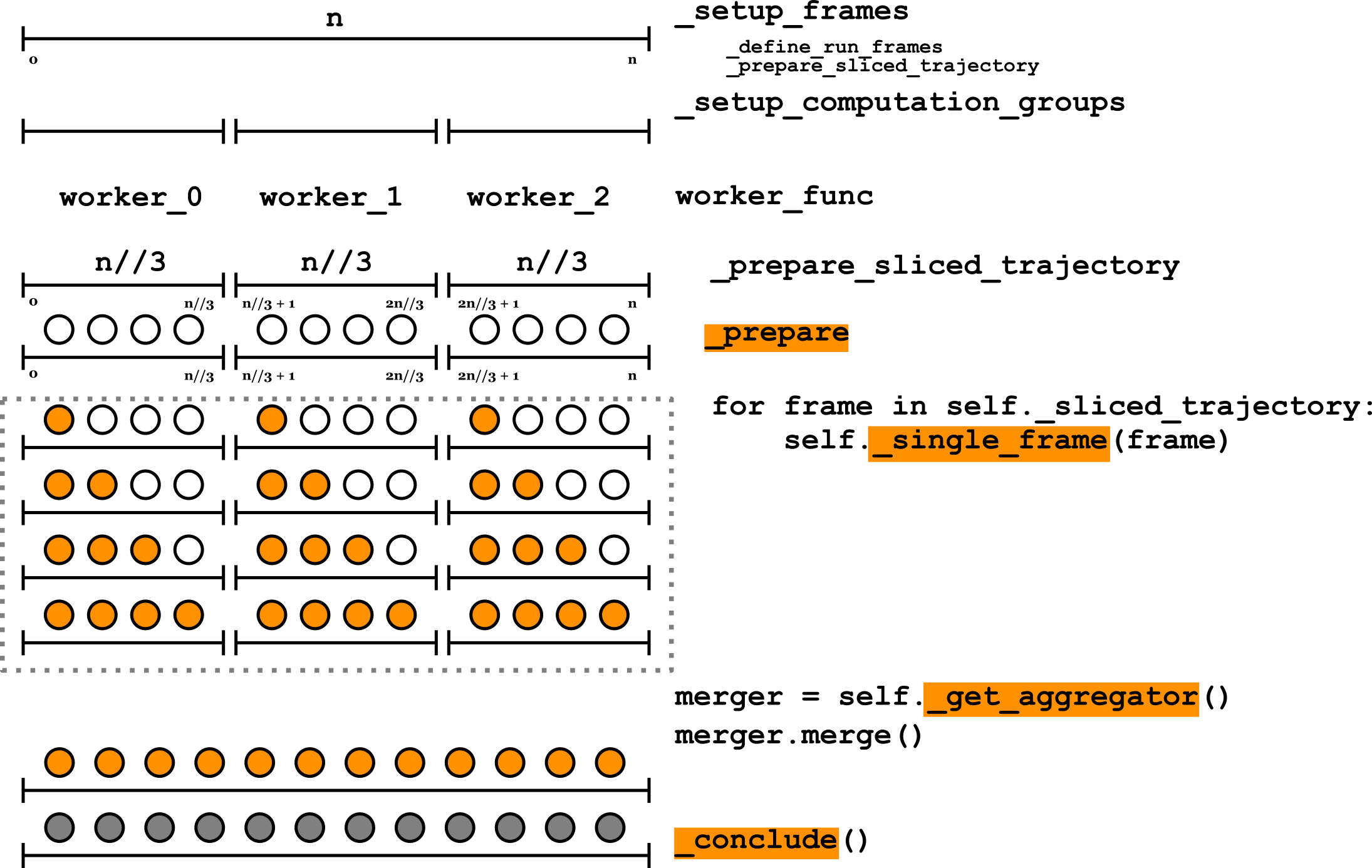
In short, after checking input parameters and configuring the backend,
AnalysisBase splits all the
frames into computation groups (equally sized sequential groups of frames to
be processed by each worker). All groups then get split between workers of
a backend configured early, the main instance gets serialized and distributed
between workers, and then
_compute() method gets called
for all frames of a computation group. Within this method, a user-implemented
_single_frame() method gets
applied to each frame in a computation group. After that, the main
instance gets an object that will combine all the objects from other
workers, and all instances get merged with an instance of
MDAnalysis.analysis.results.ResultsGroup. Then, a normal
user-implemented _compute() method
is called.
Parallelization is fully compatible with existing code and does not break
any existing code pre-2.8.0. The parallelization protocol mimics the
single-process workflow where possible. Thus, user-implemented methods such as
_prepare(),
_single_frame() and
_conclude() won’t need to know
they are operating on an instance within the main python process, or on a
remote instance, since the executed code is the same in both cases.
4.2.4.2.2. Methods in AnalysisBase for parallelization
4.2.4.2.2.1. For developers of new analysis tools
If you want to write your own parallel analysis class, you have to implement
_prepare(),
_single_frame() and
_conclude(). You also have to
denote if your analysis can run in parallel by following the steps under
Adding parallelization to your own analysis class.
Note
Attributes that are the same for the whole trajectory, should be defined in __init__ method because they need to be consistent across all workers.
4.2.4.2.2.2. For MDAnalysis developers
From a developer point of view, there are a few methods that are important in order to understand how parallelization is implemented:
MDAnalysis.analysis.base.AnalysisBase._prepare_sliced_trajectory()MDAnalysis.analysis.base.AnalysisBase._setup_computation_groups()
The first two methods share the functionality of _setup_frames().
_define_run_frames() is run once during analysis, as it checks that input
parameters start, stop, step or frames are consistent with the given
trajectory and prepares the slicer object that defines the iteration
pattern through the trajectory. _prepare_sliced_trajectory() assigns to
the self._sliced_trajectory attribute, computes the number of frames in
it, and fills the self.frames and self.times arrays. In case
the computation will be later split between other processes, this method will
be called again on each of the computation groups.
The method _configure_backend() performs basic health checks for a given
analysis class – namely, it compares a given backend (if it’s a str
instance, such as 'multiprocessing') with the list of builtin backends (and
also the backends implemented for a given analysis subclass), and configures a
MDAnalysis.analysis.backends.BackendBase instance accordingly. If the
user decides to provide a custom backend (any subclass of
MDAnalysis.analysis.backends.BackendBase, or anything with an
apply() method), it ensures that the number of workers wasn’t specified
twice (during backend initialization and in run() arguments).
After a backend is configured, _setup_computation_groups() splits the
frames prepared earlier in self._prepare_sliced_trajectory into a
number of groups, by default equal to the number of workers.
In the _compute() method, frames get initialized again with
_prepare_sliced_trajectory(), and attributes necessary for a specific
analysis get initialized with the _prepare() method. Then the function
iterates over self._sliced_trajectory, assigning
self._frame_index and self._ts as frame index (within a
computation group) and timestamp, and also setting respective
self.frames and self.times array values.
After _compute() has finished, the main analysis instance calls the
_get_aggregator() method, which merges the self.results
attributes from other processes into a single
MDAnalysis.analysis.results.Results instance, making it look for the
subsequent _conclude() method as if the run was performed in a serial
fashion, without parallelization.
4.2.4.3. Helper classes for parallelization
4.2.4.3.1. ResultsGroup
MDAnalysis.analysis.results.ResultsGroup extends the functionality of
the MDAnalysis.analysis.results.Results class. Since the Results
class is basically a dictionary that also keeps track of assigned attributes, it
is possible to iterate over all these attributes later. ResultsGroup does
exactly that: given a list of the Results objects with the same attributes,
it applies a specific aggregation function to every attribute, and stores it as
a same attribute of the returned object:
from MDAnalysis.analysis.results import ResultsGroup, Results
group = ResultsGroup(lookup={'mass': ResultsGroup.float_mean})
obj1 = Results(mass=1)
obj2 = Results(mass=3)
assert group.merge([obj1, obj2]) == Results(mass=2.0)
4.2.4.3.2. BackendBase
MDAnalysis.analysis.backends.BackendBase holds all backend attributes,
and also implements an MDAnalysis.analysis.backends.BackendBase.apply()
method, applying a given function to a list of its parameters, but in a parallel
fashion. Although in AnalysisBase it is used to apply a _compute
function, in principle it can be used to any arbitrary function and arguments,
given they’re serializable.
4.2.4.4. When to use parallelization? (Known limitations)
For now, the syntax for running parallel analysis is explicit, meaning by
default the serial version will be run, and the parallelization won’t be
enabled by default. Although we expect the parallelization to be useful in most
cases, there are some known caveats from the inital benchmarks.
4.2.4.4.1. Fast _single_frame compared to reading from disk
In all cases, parallelization will not be useful only when frames are being processed faster than being read from the disk, otherwise reading is the bottleneck here. Hence, you’ll benefit from parallelization only if you have relatively much compute per frame, or a fast drive, as illustrated below:
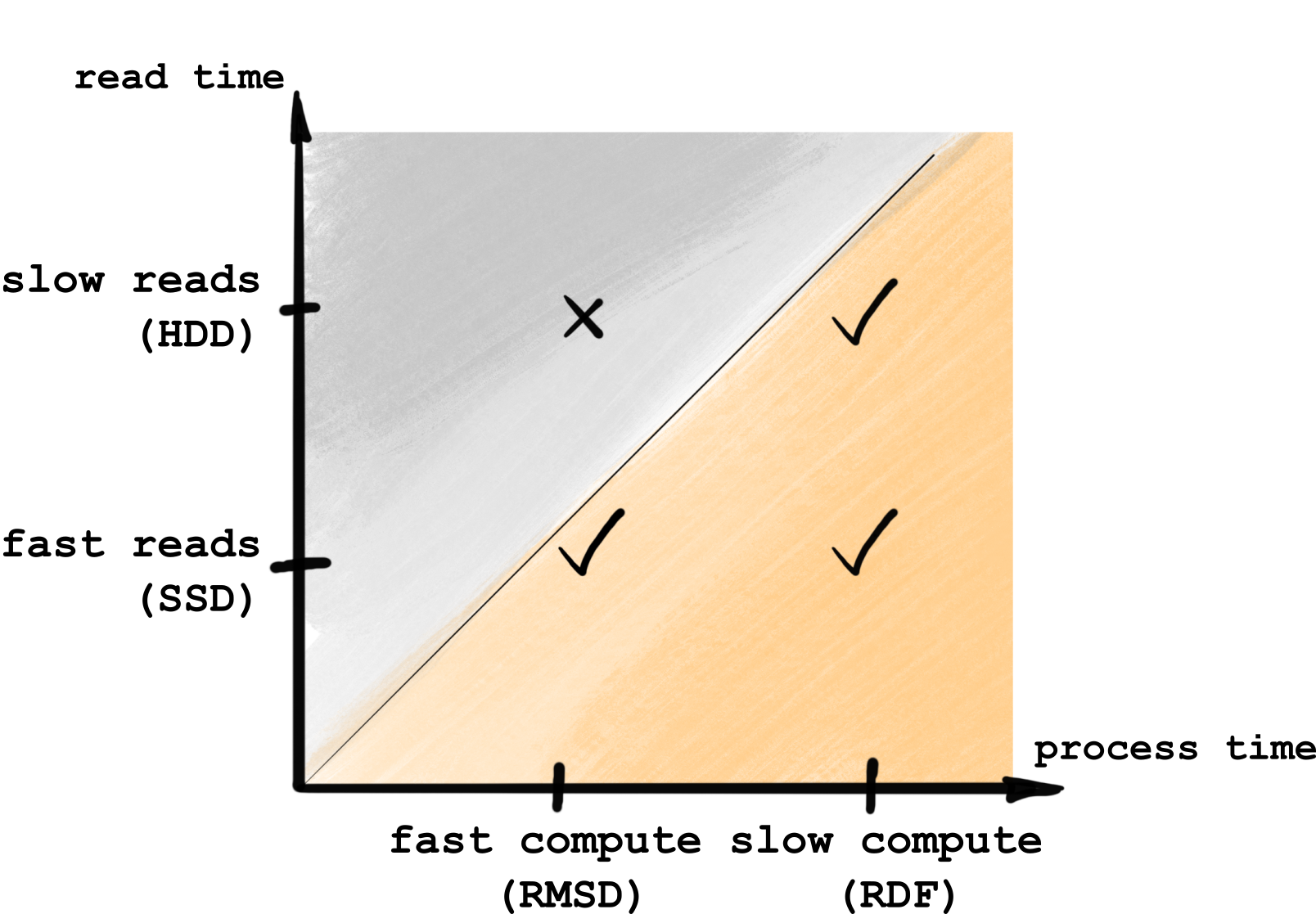
In other words, if you have fast analysis (say,
MDAnalysis.analysis.rms.RMSD) and a slow HDD drive, you are likely
to not get any benefits from parallelization. Otherwise, you should be fine.
4.2.4.4.2. Serialization issues
For built-in analysis classes, the default serialization with both
multiprocessing and dask is known to work. If you’re using some custom
analysis class that e.g. stores a non-serializable object in one of its
attributes, you might get a serialization error (PicklingError if you’re
using a multiprocessing backend). If you want to get around that, we suggest
trying backend='dask' (it uses dask serialization engine instead of
pickle).
4.2.4.4.3. Out of memory issues
If you have large memory footprint of each worker, you can run into
out-of-memory errors (i.e. your server freezes when executing a run). In this
case we suggest decreasing the number of workers from all available CPUs (that
you can get with multiprocessing.cpu_count()) to a smaller number.
4.2.4.4.4. Progress bar is missing
It is yet not possible to get a progress bar running with any parallel backend.
If you want an ETA of your analysis, we suggest running it in serial mode
for the first 10-100 frames with verbose=True, and then running it with
multiple workers. Processing time scales almost linearly, so you can get your
ETA by dividing serial ETA by the number of workers.
4.2.4.5. Adding parallelization to your own analysis class
If you want to add parallelization to your own analysis class, first make sure
your algorithm allows you to do that, i.e. you can process each frame independently.
Then it’s rather simple – let’s look at the actual code that added
parallelization to the MDAnalysis.analysis.rms.RMSD:
from MDAnalysis.analysis.base import AnalysisBase
from MDAnalysis.analysis.results import ResultsGroup
class RMSD(BackendBase):
@classmethod
def get_supported_backends(cls):
return ('serial', 'multiprocessing', 'dask',)
_analysis_algorithm_is_parallelizable = True
def _get_aggregator(self):
return ResultsGroup(lookup={'rmsd': ResultsGroup.ndarray_vstack})
That’s it! The first two methods are boilerplate –
get_supported_backends() returns a tuple with built-in backends that will
work for your class (if there are no serialization issues, it should be all
three), and _is_parallelizable is True (which is set to False in
AnalysisBase, hence we have to re-define it), and _get_aggregator()
will be used as described earlier. Note that MDAnalysis.analysis.results
also provides a few convenient functions (defined as class methods of
ResultsGroup) for results aggregation:
So you’ll likely find appropriate functions for basic aggregation there.
4.2.4.6. Writing custom backends
In order to write your custom backend (e.g. using dask.distributed), inherit
from the MDAnalysis.analysis.backends.BackendBase and (re)-implement
__init__() and apply() methods. Optionally, you can implement methods for
validation of correct backend initialization – _get_checks() and
_get_warnings(), that would raise an exception or give a warning, respectively,
when a new class instance is created:
MDAnalysis.analysis.backends._get_checks()MDAnalysis.analysis.backends._get_warnings()
from MDAnalysis.analysis.backends import BackendBase
class ThreadsBackend(BackendBase):
def __init__(self, n_workers: int, starting_message: str = "Useless backend"):
self.n_workers = n_workers
self.starting_message = starting_message
self._validate()
def _get_warnings(self):
return {True: 'warning: this backend is useless'}
def _get_checks(self):
return {isinstance(self.n_workers, int), 'error: self.n_workers is not an integer'}
def apply(self, func, computations):
from multiprocessing.dummy import Pool
with Pool(processes=self.n_workers) as pool:
print(self.starting_message)
results = pool.map(func, computations)
return results
In order to use a custom backend with another analysis class that does not
explicitly support it, you must explicitly state that you’re about to use an
unsupported_backend by passing the keyword argument
unsupported_backend=True:
from MDAnalysis.analysis.rms import RMSD
R = RMSD(...) # setup the run
n_workers = 2
backend = ThreadsBackend(n_workers=n_workers)
R.run(backend=backend, unsupported_backend=True)
In this way, you will override the check for supported backends.
Warning
When you use unsupported_backend=True you should make sure that you get
the same results as when using a supported backend for which the analysis
class was tested.
Before reporting a problem with an analysis class, make sure you tested it
with a supported backend. When reporting always mention if you used
unsupported_backend=True.
References